
About Me
I'm a passionate researcher who graduated from Texas Christian University with a B.S in Physics in May 2025. I've been actively involved in a variety of project, including computational, solid state, and atomic physics.
Currently, I work as a research associate where I combine topology and Graph Neural Networks to simulate biological phenomena. I'm developing a framework to run agent-based models to run on NVIDIA GPU's using CUDA. You can find some of my work and updates below!
My previous research endeavors include working as an undergraduate research assistant at Texas Christian University, where I studied surface properties of nanocrystalline oxides through spectroscopy, while also engineering UHV components to enable cathodoluminescence imaging. I have presented my research at various APS conferences around Texas and won multiple best presenter awards.
I'm actively searching for opportunities to apply and grow my skills. Feel free to reach out to my email or connect with me on Linkedin!
Projects
Featured
Towards a Framework For Self-Supervised Graph-Based Machine Learning
Data represented in non-euclidean space in the form of graphs provide invaluable insight into objects and their relationships with neighbours. We start with an assay of unfused cells which forms our initial graph--the nuclei are our nodes. Subsequent edges are formed by implementing the delaunay triangulation algorithm on the assay.
Graph Convolutional Networks are used to analyze the topological properties of this graph, enabling the prediction of spatial patterns such as syncytial-dominated or unfused-dominated regions. By leveraging self-supervised learning on small subgraphs--given that our nodes carry no labels--and higher-order neighbor aggregation, these networks aim to uncover consistent structural features, offering a deeper understanding of spatial heterogeneity.
My work will continue to explore the integration of persistent homology and advanced GCN architectures to enhance the detection of spatial dynamics in biological systems. Future efforts will focus on validating these models with Agent-Based Simulations (you can find a simple simulation here) and expanding to diverse datasets, aiming to contribute meaningful insights to graph-based machine learning.
View Source Code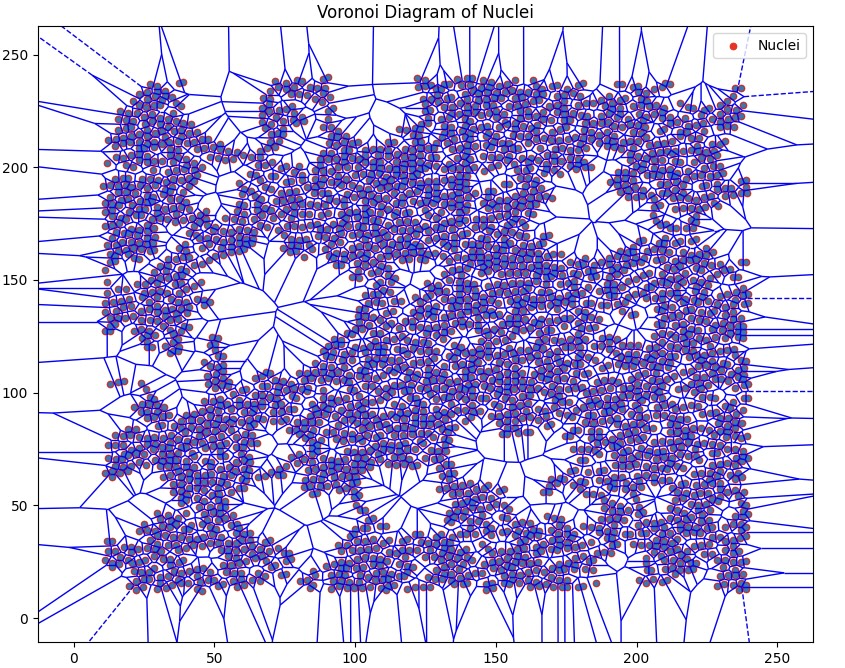
Voronoi diagram
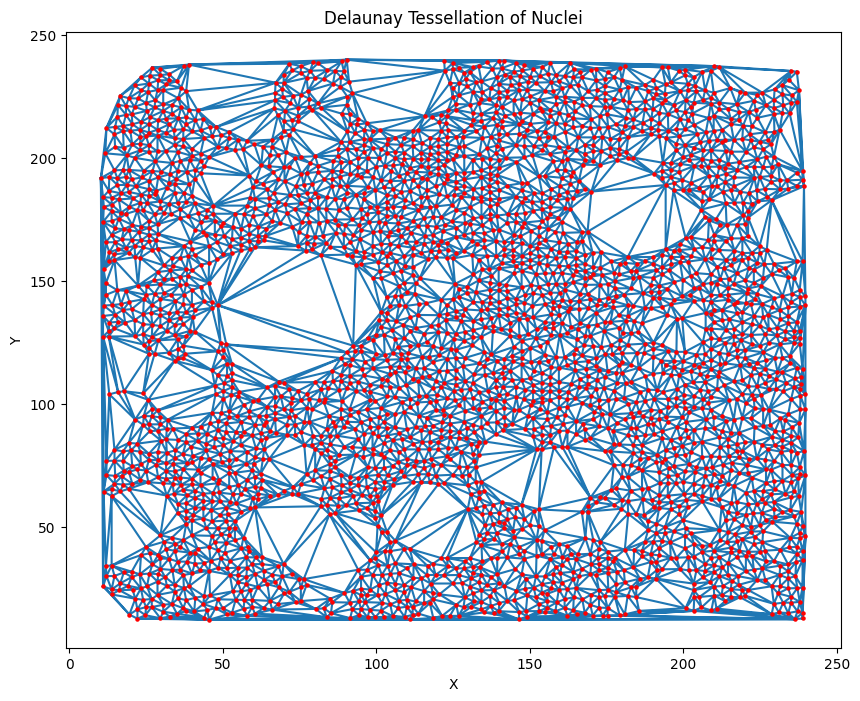
Delaunay triangulation
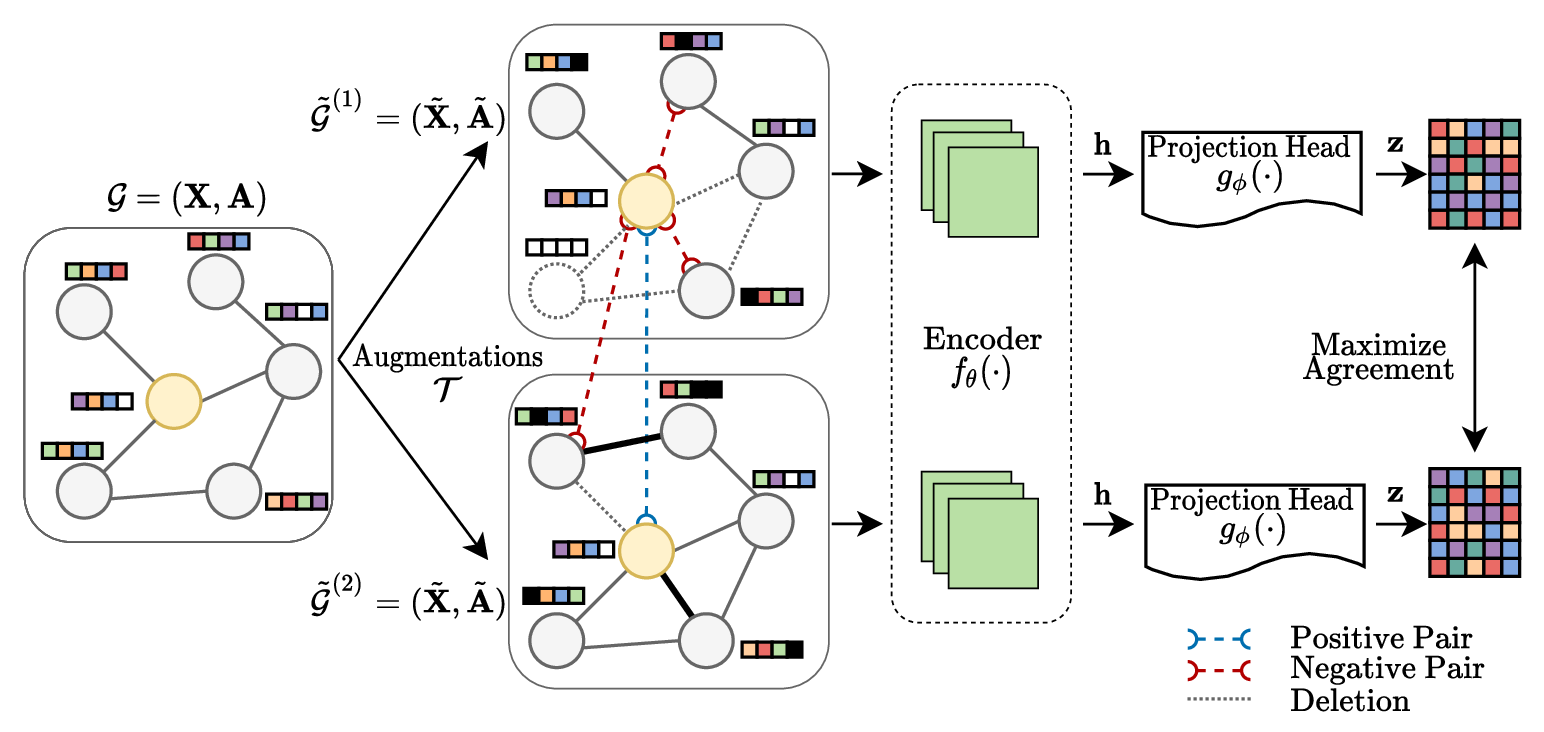
Graph Contrastive Learning
Other Projects
Topological Data Analysis
Using persistence homology to evaluate data
Agent-Based Model
Web-based simulation of cell-cell fusion, utilising Flask for backend and allowing users to adjust parameters.
Pandemic Simulation
Tuneable Python script that uses physical perturbation to model human movement and track virus spread in a pandemic.